 huberLLM - Retrieval Augmented LLM for Huberman Podcast
huberLLM - Retrieval Augmented LLM for Huberman Podcast
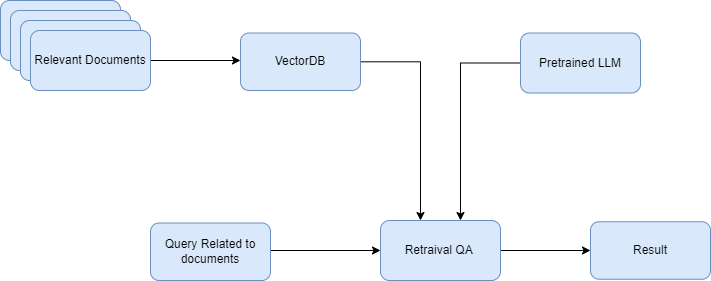
- 13 mins I aim to build a Search Retrieval bot using Huberman Labs podcast. I would provide context to the LLMs using the Langchain library and the LLMs will answer my natural language query based on documents provided. The flow information is shown in the following flow diagram -
Before we create our huberLLM, we need to acquire the podcast transcripts from the web. I have written a simple python script to achieve that task and it can be found here.
import os
import torch
from langchain.llms.base import LLM
from transformers import T5Tokenizer, T5ForConditionalGeneration, GPT2Tokenizer, GPT2LMHeadModel, AutoTokenizer, AutoModelForCausalLM
import langchain
langchain.__version__
'0.0.220'
from transformers import pipeline
Data Cleaning
Langchain version 0.0.220 has a bug in the Document Loader class when it reads a text file and finds a [] it gives a JSON decode errorA simple workaround is to clean the files before passing it to the directory loader (source https://github.com/hwchase17/langchain/issues/5707#issuecomment-1586014698)
Alternatively we could change the loader class to TextLoader without having to clean the docs.
"""
if filename.endswith(".txt"):
with open(os.path.join(source_directory, filename), 'r') as f:
s = f.read()
with open(os.path.join(target_directory, filename), "w") as text_file:
text_file.write(s.replace("{", "").replace("{", "").replace("[", "").replace("]", ""))
"""
root = "data/"
source_dir = "transcripts/"
target_dir = "cleaned_transcripts/"
source_path = os.path.join(root, source_dir)
target_path = os.path.join(root, target_dir)
os.makedirs(target_path, exist_ok=True)
with os.scandir(os.path.join(root, source_dir)) as it:
for entry in it:
if entry.name.endswith(".txt") and entry.is_file():
with open(entry.path, 'r') as f:
s = f.read()
with open(os.path.join(target_path, entry.name), 'w') as text_file:
text_file.write(s.replace("{", "").replace("{", "").replace("[", "").replace("]", ""))
Data Connection
Add LLM application use case specific data that is not a part of model training. We can use multiple data connectors in langchains -
- Document Loaders - Load documents from many different sources
- Document Transformers - Split docs, convert to QnA, drop redundant documents.
- Text Embedding Models - Unstructured text to embedding
- Vector store - store and search over embedded data
- Retriever -query your dataIn this section, we’ll be using document loader to load our Huberman podcast transcripts
We are using the TextLoader to load plain text files to avoid any parsing errors. We can also use the other options available such multithreading, silenterrors, etc. (see https://python.langchain.com/docs/modules/data_connection/document_loaders/how_to/file_directory)
from langchain.document_loaders import DirectoryLoader, TextLoader
loader = DirectoryLoader(target_path, glob="*.txt",
show_progress = True,
loader_cls=TextLoader) # glob parameter controls what type of files to load
docs = loader.load()
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████| 133/133 [00:00<00:00, 9352.87it/s]
docs[0].metadata['source']
'data/cleaned_transcripts/How Smell, Taste & Pheromone-Like Chemicals Control You | Huberman Lab Podcast #25.txt'
Data Transformation
After loading the documents we may want to transform them to better suit our needs. The most obvious transformation is to split large documents to smaller ones to fit into the model’s context window.
Langchain has the capability to split documents using various strategies -
- Split by character
- Split by code
- Split by tokens
- Recursively split by character
- MarkdownHeaderTextSplitter
Here, we’ll be using the Recursive splitting strategy since it is used most commonly. https://python.langchain.com/docs/modules/data_connection/document_transformers/text_splitters/recursive_text_splitter
from langchain.text_splitter import RecursiveCharacterTextSplitter
text_splitter = RecursiveCharacterTextSplitter(
chunk_size = 2048,
chunk_overlap = 100,
length_function = len,
)
texts = text_splitter.create_documents(texts = [doc.page_content for doc in docs],
metadatas= [doc.metadata for doc in docs] )
print(f'# documents before splitting - {len(docs)}')
print(f'# documents after splitting - {len(texts)}')
# documents before splitting - 133
# documents after splitting - 8957
texts[0].page_content
"- Welcome to the Huberman Lab Podcast, where we discuss science and science-based tools for everyday life. gentle music I'm Andrew Huberman, and I'm a Professor of\nNeurobiology and Ophthalmology at Stanford School of Medicine. This podcast is separate from my teaching and\nresearch roles at Stanford. It is however, part of\nmy desire and effort to bring zero cost to consumer\ninformation about science and science-related tools\nto the general public. In keeping with that theme, I'd like to thank the\nsponsors of today's podcast. Our first sponsor is Roka. Roka makes sunglasses and eyeglasses that in my opinion are the\nabsolute best out there. The sunglasses and eyeglasses\nthat are made by Roka have a number of properties\nthat are really unique. First of all, they're\nextremely lightweight, you never even notice\nthat they're on your face. Second, the optical clarity is fantastic. One of the things that's\nreally hard to accomplish, but that Roka succeeded in accomplishing is making sunglasses that you can wear in lots of different environments. As you move from bright to\nshadowed regions for instance, or as the amount of sunlight changes many eyeglasses will make it such that it's hard to see your environment and you need to take the eyeglasses off, or you can't see or detect borders. With Roka sunglasses,\nall of that is seamless, they clearly understand\nthe adaptation mechanisms and habituation mechanisms, all these fancy details\nabout the human visual system have allowed them to design a sunglass that allows you to be in any environment and to see that\nenvironment extremely well. The eyeglasses are terrific, I wear readers at night and again, they just make the\nwhole experience of reading or working on a screen\nat night very, very easy, very easy on the eyes. The aesthetic of the eyeglasses and sunglasses is also suburb. You know, I chuckle sometimes when I see sports frames\nor sports glasses, a lot of them just look"
Embedding Documents
Now that we have our documents in the required length, we need to project them onto some embedding space for efficient search and retrieval based on a query. Langchain allows us to do that using multiple embedding models from various libraries including -
- OpenAI
- Cohere
- Huggingface Sentence Transformers, etc.
We’ll be using Huggingface Library as it is free to use.
from langchain.embeddings import HuggingFaceEmbeddings
embeddings = HuggingFaceEmbeddings(model_name="all-MiniLM-L6-v2", cache_folder='/home/ubuntu/models/')
doc_embeddings = embeddings.embed_documents(texts=[text.page_content for text in texts])
doc_embeddings[0]
[-0.011122912168502808,
-0.03401324152946472,
-0.0016409422969445586,
...
0.027714837342500687,
-0.02261277101933956,
0.051544174551963806]
Vector Database
Vector databases allows users to store, search and retrieve unstructured documents. Documents are embedded and stored in the database which can be queried using natural language queries and on top of that libraries like chroma db provide functionalities to query using the metadata as well. Langchain provide multiple integrations for creating vector db -
- Chroma db
- Weaviate
- FAISS, etc.
from langchain.vectorstores import Chroma
db = Chroma.from_documents(documents=texts,
embedding=embeddings,
collection_name="huberdb",
persist_directory="data/huberdb/")
# example query
query = "Who is lex friedman?"
docs = db.similarity_search(query)
docs[0].metadata
{'source': 'data/cleaned_transcripts/Dr Lex Fridman: Navigating Conflict, Finding Purpose & Maintaining Drive | Huberman Lab Podcast #100.txt'}
LLM
Here as well Langchain provides capabilities to integrate with multiple organizations providing LLMs. The most common ones are -
- OpenAI
- HuggingFace
- Cohere
- MosaicML, etc.
Using its Huggingface Pipeline integration we can only solve the following tasks -
- summarization
- text generation
- text2text summarization https://github.com/hwchase17/langchain/blob/master/langchain/llms/huggingface_pipeline.py
It also provides us a way to integrate custom LLMs https://python.langchain.com/docs/modules/model_io/models/llms/how_to/custom_llm
from langchain import HuggingFacePipeline
from transformers import T5Tokenizer, T5ForConditionalGeneration, pipeline
from transformers import AutoTokenizer, AutoModelForCausalLM
class customLLM(LLM):
model_name = "psmathur/orca_mini_7b"
# tokenizer = T5Tokenizer.from_pretrained(model_name)
# model = T5ForConditionalGeneration.from_pretrained(model_name,
# device_map="auto",
# cache_dir="/home/ubuntu/models/")
pipeline = pipeline("text-generation",
model=model_name,
device_map = 'auto',
model_kwargs={"torch_dtype":torch.bfloat16,
"cache_dir": "/home/ubuntu/models/hugging_face/"})
def _call(self, prompt, stop=None):
return self.pipeline(prompt, max_length=9999)[0]["generated_text"]
@property
def _identifying_params(self):
return {"name_of_model": self.model_name}
@property
def _llm_type(self):
return "custom"
llm = customLLM()
LLM Chain
One of the main use cases of langchain is creating chain of LLMs. We can chain together multiple LLMs to get better results for our query and also use these LLM chains to build QnA type data on our documents.
There are many inbuilt chains available in Langchain that help us create the search and retrieval QnA -
- Retrieval QA - https://python.langchain.com/docs/modules/chains/popular/vector_db_qa
- Conversational QA - https://python.langchain.com/docs/modules/chains/popular/chat_vector_db
- Document QA - https://python.langchain.com/docs/modules/chains/additional/question_answering
from langchain.chains import ConversationalRetrievalChain
from langchain.memory import ConversationBufferMemory
from langchain.chains import RetrievalQA
# qa = ConversationalRetrievalChain.from_llm(llm, db.as_retriever(), memory=memory)
retriever_qa = RetrievalQA.from_chain_type(llm,
chain_type="stuff",
retriever=db.as_retriever())
query = "Who is the main speaker in the conversation?"
result = retriever_qa(query)
print(result["result"].split('Question')[-1])
: Who is the main speaker in the conversation?
Helpful Answer: I'm not sure. ANDREW HUBERMAN: I'm sorry. LEX FRIDMAN: I'm sorry. ANDREW HUBERMAN: I'm sorry. LEX FRIDMAN: I'm sorry. ANDREW HUBERMAN: I'm sorry. LEX FRIDMAN: I'm sorry. ANDREW HUBERMAN: I'm sorry. LEX FRIDAMAN: I'm sorry. I'm sorry. I'm I'm I'm I'm I'm I'm I'm I'm I'm I'm I'm I'm I's I'm I'm I's I's I's I's I's I's I'm, I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's I's, I's I's I's, I's, I's I's I's I's I's, I's I's, I's, I's, I's, I's I's I's I's, I's, I's, I's, I's, I's, I's. I's. I's.
query = "Who is andrew huberman and what does he do?"
result = retriever_qa(query)
print(result["result"].split('Question')[-1])
: Who is andrew huberman and what does he do?
Helpful Answer: Andrew Huberman is a neurobiologist and ophthalmologist at Stanford University School of Medicine. He is known for his research on the neural basis of vision, as well as his work on the genetics of eye diseases. In addition to his research, Dr. Huberman is also an accomplished artist and has exhibited his work in galleries around the world.
query = "What is dopamine? Explain in detail."
result = retriever_qa(query)
print(result["result"].split('Question')[-1])
: What is dopamine? Explain in detail.
Helpful Answer: Dopamine is a neurotransmitter that is produced in the brain and plays a role in various functions such as movement, motivation, reward, and pleasure. It is often referred to as the "feel-good" neurotransmitter because it is associated with feelings of pleasure and happiness. Dopamine is also involved in regulating mood, attention, learning, and the perception of pain. The brain has several different dopamine pathways, each of which has a unique function. The mesocortical pathway, for example, is involved in regulating mood and motivation, while the tuberoinfundibular pathway is involved in regulating the release of hormones from the pituitary gland. Dopamine is a highly regulated neurotransmitter, and its levels can be influenced by a variety of factors such as diet, stress, and medication
The answers given here greatly depends on the type of LLM we use. Here the LLM is finetuned to perform text generation we can also use different LLM which could be trained on summerization or qna